A comprehensive evolutionary and molecular modelling suite that comprises of a series of tools and workflows for drug design developed by our group.
(Download here)

Drugster is a freeware platform aimed to assist scientists in the field of Computer Aided Drug Design.
(Download here)
DrugOn is a fully interactive pipeline designed to overcome the command line barrier with two friendly environments for the user.
(Download here)
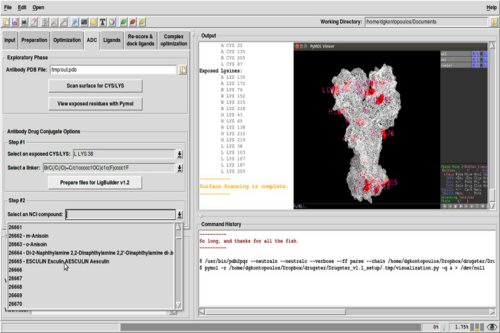
Drugena encompasses an up-to-date database of antibodies for neurological disorders and the NCI database for the in silico development of ADCs.
(Download here)

Antisoma application has been developed in order to provide the information found with this new approach and concerning the antibodies of the species.
(Download here)

With the extensive use of microarray technology as a potential prognostic and diagnostic tool, the comparison and reproducibility of results obtained from the use of different platforms is of interest.
(Download here)

Space Suite, a new homology modelling approach, successfully applied to the homology modelling of the RNA-dependent RNA polymerase (RdRp) of dengue (type II) virus.
(Download here)

A 'molecular cartography' suite for the two-dimensional transformation of protein structures the multidimensional analysis of their physicochemical properties.
(more info here)
(Download here)

Taggo takes advantage of the Gene Ontology (GO) to extract the proteins’ main attributes and combines the potential of discarding annotations that are supported by not so reliable Ecs.
(Download here)

The Brukin2d software, is a visualization tool for large data sets produced from LC-MS. Specifically the chromatograph and the mass spectra that correspond to its peaks is the focus of this work.
(Download here)

GIBA is an effective and easy to use tool for the detection of protein complexes through clustering PPI networks. GIBA surpasses other methods in quality approximations of protein complexes.
(Download here)

AHC is a hierarchical algorithm that performs successive min–cuts until it identifies dense subgraphs. AHC leads to the selection of more dense subgraphs as protein complexes candidates.
(Download here)

GAppi performs clustering in PPI networks to identify protein complexes. The algorithm has been tested exhaustively and results showed that GAppi outperforms other techniques producing feasible and very efficient solutions.
(Download here)

GOmir (by using up to four different databases) introduces, for the first time, miRNA predicted targets accompanied by (a) full gene description, (b) functional analysis and (c) detailed gene ontology clustering.
(Download here)

A tool that encompasses retrieval and analysis of fusion events used to identify putative protein-protein interactions in completely sequenced genomes of various prokaryotes, and eukaryotes.
(Download FED here)
(Download SAFE here)

A suite of computer programs that has been developed for monitoring structural changes on a-helical secondary structural elements during molecular dynamics (MD) simulations on protein systems.
(Download here)